Geometry and mesh using SALOME
Please read
Part I of this article.
CFD solving with Code_Saturne
After the mesh file is produced from SALOME, Code_Saturne can be used for a CFD calculation.
Note that I take Code_Saturne 2.0.0 as an example in this post; if you are still using Code_Saturne 1.x, remember some commands and directory names are different.
1. Create a study with a case.
Command 'cs create' is for this. Use option '--help' to get its help message, '-s' and '-c' to specify a study name and a case name, respectively.
Note that from 2.0-rc1 version, the script name
cs is changed to
code_saturne; please use
code_saturne instead if you are using Code_Saturne 2.0-rc1. (
Within Code_Saturne 1.x, command 'cree_sat -etude' has to be used instead; for clarity, this post doesn't include the content related to version 1.x.)
:/$ cs create --help
:/$ cs create -s DUCT -c 2D
With the command executed, a study directory named 'DUCT' and a case called '2D' are created. In the directory 'DUCT', there are three sub-directories:
*.
2D: The case directory. A study can have several cases; for example, different boundary conditions can be organised as different cases.
*.
MESH: Code_Saturne will read mesh files from this directory, and as such we need to copy mesh files into it;
*.
POST: This is an empty directory designed to contain post-processing macros (xmgrace, experimental etc).
Copy the mesh file 'mesh_d.med' into the directory 'MESH'. Then we look at the case directory '2D', in which there are four sub-directories:
*.
DATA: Contains the script to launch the GUI SaturneGUI.
*.
RESU: Directory where the results will be exported once the calculation has finished.
*.
SCRIPTS: The launching scripts are copied here. The CFD calculation is actually started by executing the launching script 'runcase' here.
*.
SRC: The user subroutines are saved here. During the compilation stage, the code will re-compile all the subroutines that are in this directory. In the 'REFERENCE' directory all the user-defined subroutines are stored. For a specific case you will only need a few of them depending on the changes that you want to implement on the code. The 'base' directory contains the basic flow solver subroutines, and the other directories contain the subroutines needed by other modules.
2. Use GUI to input parameters. Ship into the directory 'DUCT/2D/DATA' and then execute 'SaturneGUI' to launch the GUI.
:/$ ./SaturneGUI &
The parameters set with the GUI can actually be saved into a XML file, and as such for a second time to launch the GUI, option '-f' can be used to load the XML file directly.
:/$ ./SaturneGUI -f 2d_duct_flow.xml &
The first glance at the GUI is like this. It lists the structure of directories. With the help of the navigator at the left panel, we can go to different pages to modify various parameters.
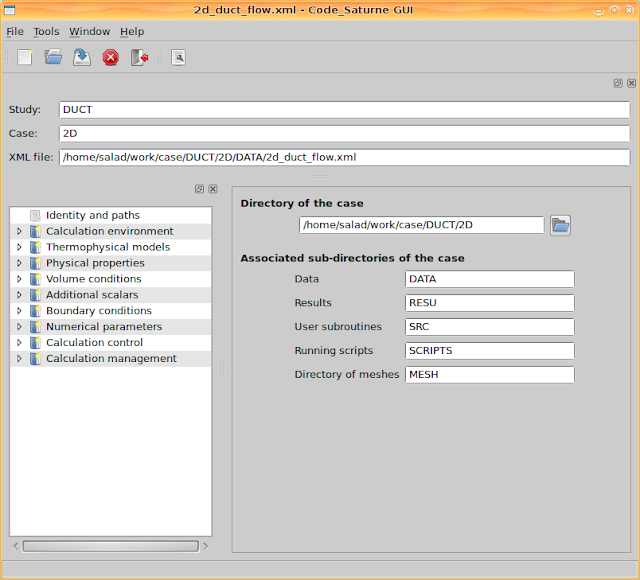
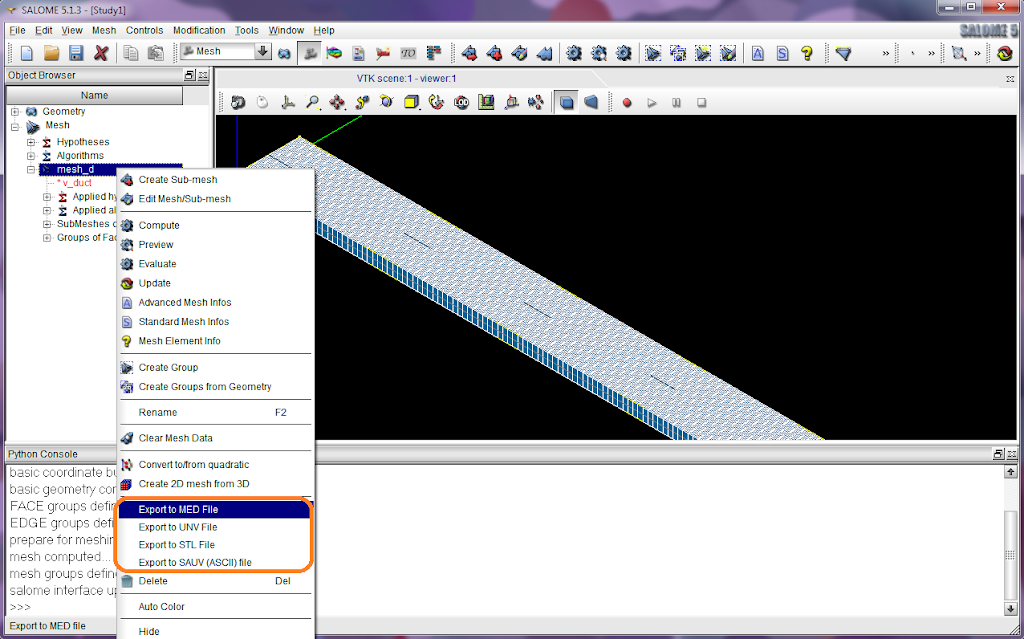
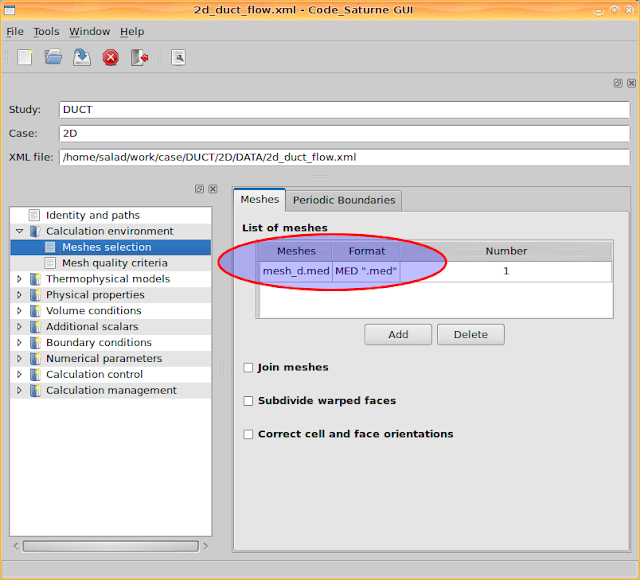
Go to 'Calculation environment > Meshes selection' and add the mesh file 'mesh_d.med'. It can recognise mesh files from the directory 'MESH'.
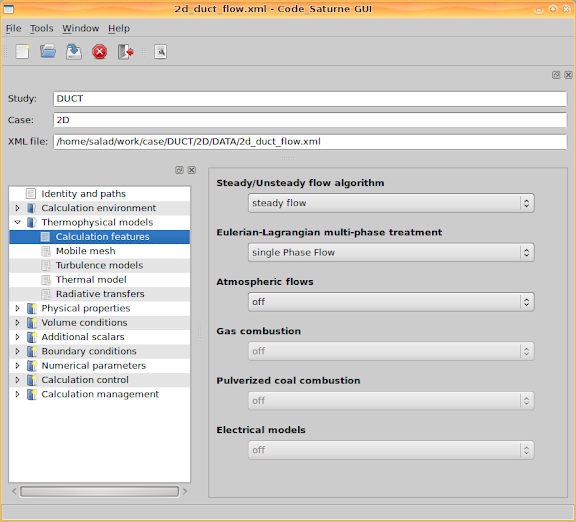
Go to 'Thermophysical models > Calculation features' and enable two features, 'steady flow' and 'single Phase Flow'.
Go to 'Thermophysical models > Turbulence models' and select 'No model (i.e. laminar flow), since we ignore turbulence and only consider a laminar approximation. Besides, select the 'Thermal scalar' to be 'Temperature (Celsius)' on the page 'Thermal model' and select 'No radiative transfers' on the page 'Radiative transfers' as we ignore radiation.
Go to 'Physical properties > Fluid properties' and set the four fluid properties: density, viscosity, specific heat and thermal conductivity. As shown in the following figure, we set specific heat and thermal conductivity as constant values, and leave density and viscosity to the user subroutine 'usphyv'. Therefore, the reference values set for density and viscosity do not matter.
In 'usphyv' the parameters could be controlled as temperature dependent. It will be discussed in the next section.

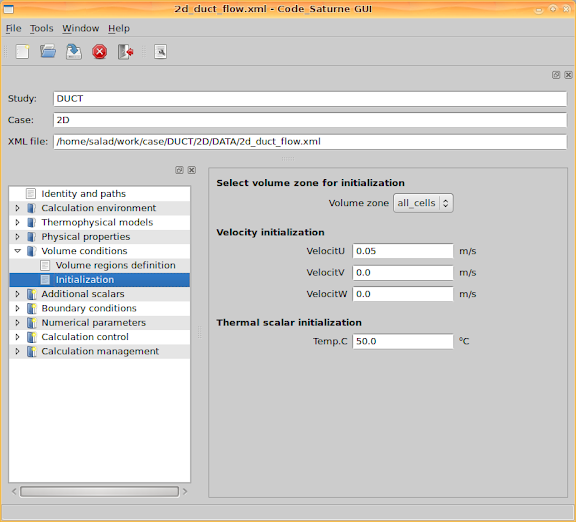
Go to 'Volume conditions > Initialization' and set initial values for velocity and temperature. Here I set the x component of the velocity as the inlet velocity value, and set the temperature as the inlet temperature value.
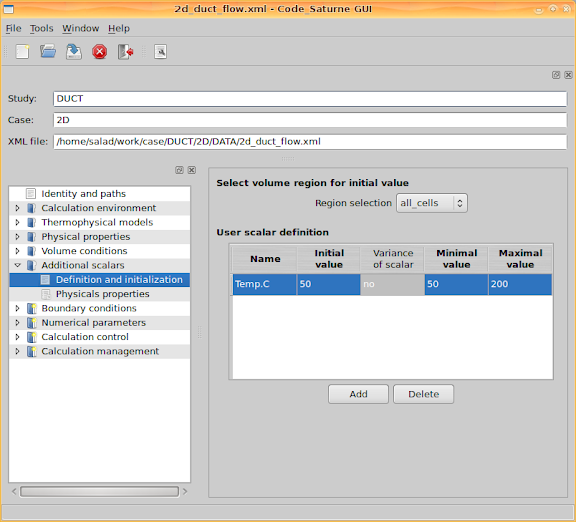
Go to 'Additional scalars > Definition and initialization' and set the scalar limitation for temperature, including its minimal and maximal values.
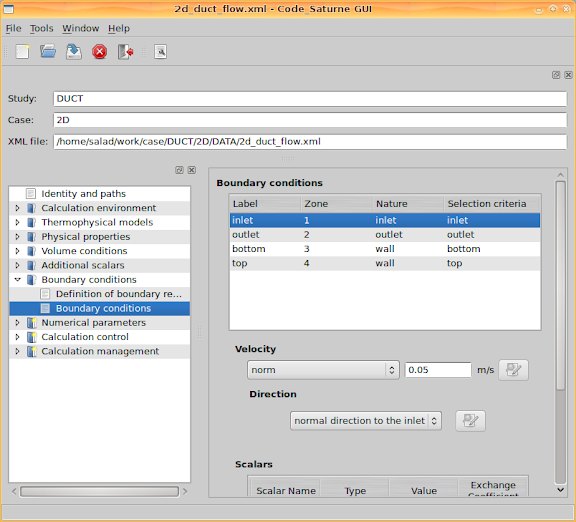
Next step is about boundary conditions. Firstly, boundaries have to be defined. Since we used SALOME to generate the mesh, according to the defined mesh groups, inlet, outlet, bottom, top and sym, corresponding boundaries can be defined. For more details about the mesh groups, please see
Part I of this article.

Secondly, go to 'Boundary conditions > Boundary conditions' and set values for the boundaries. I set the inlet velocity and temperature, and set minus heat flux values at the two walls. Negative heat flux means the heat is into the channel. Additionally, the walls are set as smooth walls.
Leave 'Numerical parameters' with the default values, and go to 'Calculation control > Output control'. Then set 'Output listing at each time step', set output 'Post-processing' 'Only at the end of calculation' since we are concerning a steady state calculation result.
With respect to the 'Post-processing format', either 'Ensight Gold' or 'MED' can be specified. If you prefer to use ParaView, select 'Ensight Gold'. If you want to use SALOME to do post-processing, change the option to 'MED'.
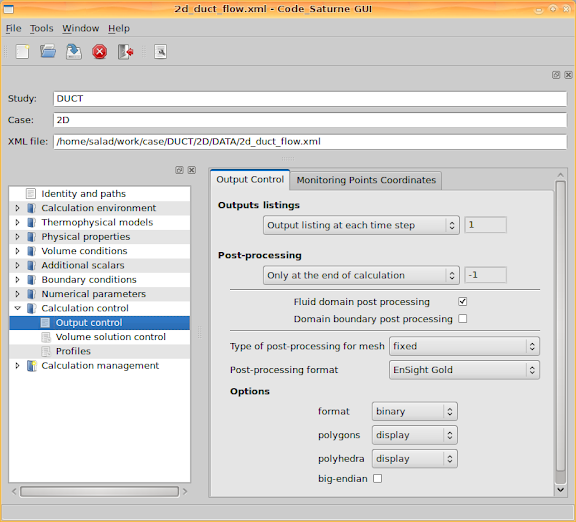
Then go to the second tab page. This page lists the 'Monitoring points coordinates'. When the calculation is being performed, the quantity values at these coordinates will be recorded for each iteration step (each time step if it is a transient process).
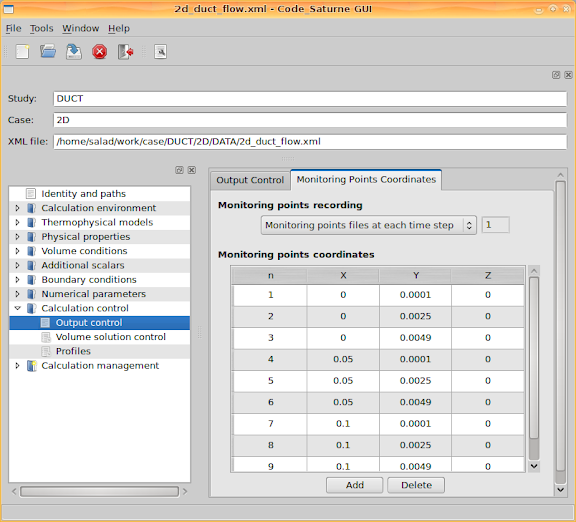
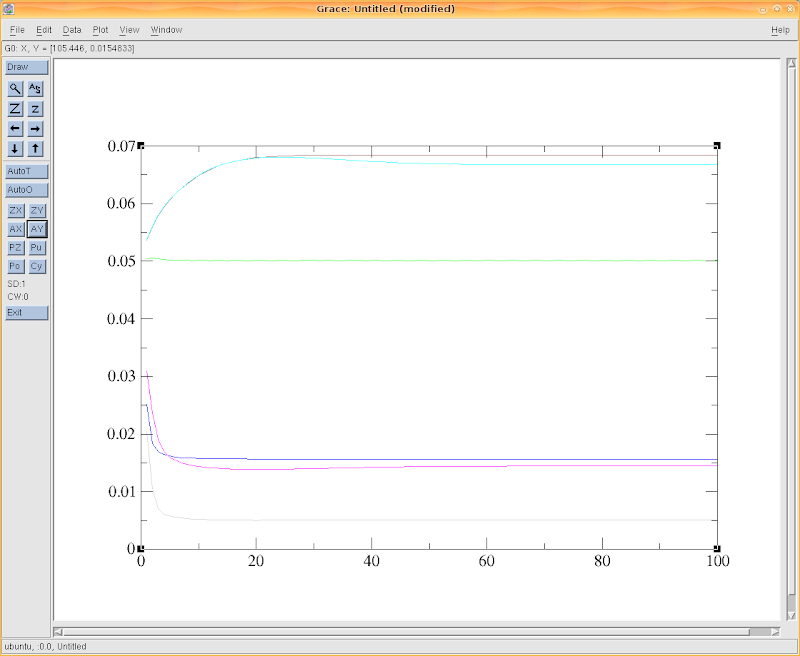
Once the calculation is started, we can go to Saturne's temporary directory to check the history data at these monitored points. For example, to check the x component of velocity, use 'xmgrace'
~/tmp_Saturne/DUCT.2D.02061151$ xmgrace -nxy probes_VelocitU.dat
The result could be like this figure below, from which we can see all the values are already stable after 70 iteration steps. It implies a criterion to stop the calculation after a number of steps.
_VelocitU.png)
Actually
cs plot_probes command instead of
xmgrace -nxy could be used to draw the graph of the values at the probes; it's a wrapper around
xmgrace with a couple of file manipulation.
3. Apply FORTRAN user routines.
As mentioned the fluid density and viscosity could be configured as temperature dependent by modifying user routines in 'usphyv.f90'. Adopting FORTRAN code to control the calculation details brings enough flexibility to Code_Saturne.
All user routines/FORTRAN files are put in the directory 'SRC'. Look up the sub-directory 'REFERENCE' and see all the routines you can modify. For fluid properties, copy the file SRC/REFERENCE/base/usphyv.f90 into SRC and then modify the copied file in SRC.
For density, search the corresponding piece of code in the file and then modify the part as the following code list, which defines the density expression
}) T
T is temperature. Actually 0.00065 is the fluid expansion coefficient due to temperature.
iutile = 1
! ...
! --- Coefficients des lois choisis et imposes par l'utilisateur
! Les valeurs donnees ici sont fictives
vara = 895
varb = 0.00065
varc = 20
! Masse volumique au centre des cellules
! ---------------------------------------
! loi RHO = A / (1 + B * (T - C))
! soit PROPCE(IEL,IPCROM) = VARA / (1 + VARB * (XRTP - VARC))
do iel = 1, ncel
xrtp = rtp(iel, ivart)
propce(iel, ipcrom) = vara / (1 + varb * (xrtp - varc))
enddo
Note that the first line set the variable 'iutile' to 1, otherwise the code implementing the equation will not take effect.
For viscosity, find the code piece, assign 'iutile' 1 and modify the following code as whatever viscosity equation you would try. With the density code example, it is not supposed to be difficult.
!===============================================================================
! EXEMPLE 2 : VISCOSITE VARIABLE EN FONCTION DE LA TEMPERATURE
! ===========
! ...
iutile = 1
! ...
4. Solve.
After configured all the necessary aspects and modified the user routines, go to 'Calculation management > Prepare batch calculation' and we are able to start the CFD calculation right now. Firstly, 'Select the batch script file', i.e. click the button to browser a launch script file. As default, the file is named 'runcase'.
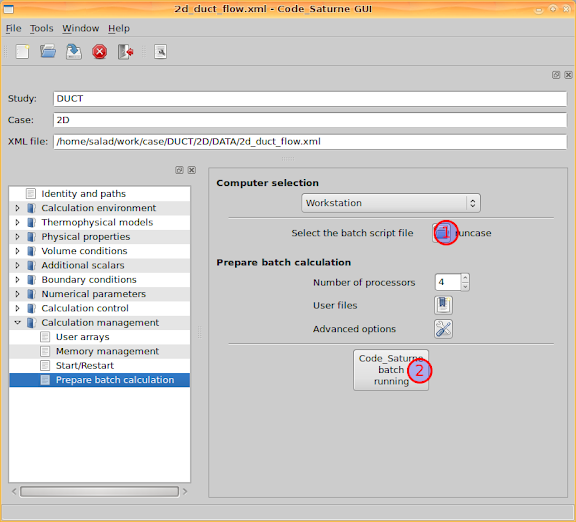
Then select the 'Number of processors', and click the button 'Code_Saturne batch running'. A console window will pop out and Code_Saturne will produce code to do the calculation. I assume that you are using more than one processor cores to calculate, and then you might be asked for your Linux system user password by openmpi before the calculation.
Leave the calculation console for minutes and then, if lucky enough, you will see the calculation finished normally. If so, the next step is to see the results and do post-processing.
Post-processing with ParaView
Please read
Part III of this article.











_VelocitU.png)